Keras
May 4, 2023
June 29, 2019
Model accuracy is not a reliable metric of performance, because it will yield misleading results if the validation data set is unbalanced. For example, if there were 90 cats and only 10 dogs in the validation data set and if the model predicts all the images as cats. The overall accuracy would be 90%.
The confusion matrix allows us to visualize the performance of the trained model. It makes it easy to see if the system is confusing two classes. It also summarizes the results of testing the model for further inspection. In this tutorial, we create a simple Convolutional Neural Network (CNN) to classify MNIST digits for visualization confusion matrix in TensorBord.
Download Dataset
We’re going to construct a simple neural network to classify images in the MNIST dataset. This dataset consists of 28×28 grayscale images of 10 digits(0-9) of 10 categories.
(train_images, train_labels), (test_images, test_labels) = datasets.mnist.load_data() train_images = train_images.reshape((60000, 28, 28, 1)) test_images = test_images.reshape((10000, 28, 28, 1)) train_images, test_images = train_images / 255.0, test_images / 255.0 classes=[0,1,2,3,4,5,6,7,8,9]
Define Simple CNN Model
First, create a very simple model and compile it, setting up the optimizer and loss function and training it.
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(28, 28, 1)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.Flatten())
model.add(layers.Dense(64, activation='relu'))
model.add(layers.Dense(10, activation='softmax'))
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
model.fit(x=train_images,
y=train_labels,
epochs=5,
validation_data=(test_images, test_labels))
The compile step also specifies that you want to log the accuracy of the classifier along the way.
Create a Confusion Matrix
You can use Tensorflow’s confusion matrix to create a confusion matrix.
y_pred=model.predict_classes(test_images) con_mat = tf.math.confusion_matrix(labels=y_true, predictions=y_pred).numpy()
Normalization Confusion Matrix to the interpretation of which class is being misclassified.
con_mat_norm = np.around(con_mat.astype('float') / con_mat.sum(axis=1)[:, np.newaxis], decimals=2)
con_mat_df = pd.DataFrame(con_mat_norm,
index = classes,
columns = classes)

The diagonal elements represent the number of points for which the predicted label is equal to the true label, while off-diagonal elements are those that are mislabeled by the model.
figure = plt.figure(figsize=(8, 8))
sns.heatmap(con_mat_df, annot=True,cmap=plt.cm.Blues)
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt.show()
We use matplotlib to plot confusion matrix and Seaborn library to create a heatmap.

The confusion matrix shows that this model has some problems. “9”, “5”, and “2” are getting confused with each other. The model needs more work.
Plot Confusion Matrix in Tensorbord
Using the TensorFlow Image Summary API, you can easily view them in TensorBoard.Here’s what you’ll do:
- Create the Keras TensorBoard callback to log basic metrics
- Create a Keras LambdaCallback to log the confusion matrix at the end of every epoch
- Train the model using Model.fit(), making sure to pass both callbacks
You need some boilerplate code to convert the plot to a tensor, tf.summary.image() expecting a rank-4 tensor containing (batch_size, height, width, and channels). Therefore, the tensors need to be reshaped.
file_writer = tf.summary.create_file_writer(logdir + '/cm')
def log_confusion_matrix(epoch, logs):
# Use the model to predict the values from the validation dataset.
test_pred = model1.predict_classes(test_images)
con_mat = tf.math.confusion_matrix(labels=test_labels, predictions=test_pred).numpy()
con_mat_norm = np.around(con_mat.astype('float') / con_mat.sum(axis=1)[:, np.newaxis], decimals=2)
con_mat_df = pd.DataFrame(con_mat_norm,
index = classes,
columns = classes)
figure = plt.figure(figsize=(8, 8))
sns.heatmap(con_mat_df, annot=True,cmap=plt.cm.Blues)
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
buf = io.BytesIO()
plt.savefig(buf, format='png')
plt.close(figure)
buf.seek(0)
image = tf.image.decode_png(buf.getvalue(), channels=4)
image = tf.expand_dims(image, 0)
# Log the confusion matrix as an image summary.
with file_writer.as_default():
tf.summary.image("Confusion Matrix", image, step=epoch)
logdir='logs/images'
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=logdir)
cm_callback = keras.callbacks.LambdaCallback(on_epoch_end=log_confusion_matrix)
You’re now ready to train the model and log this image and view it in TensorBoard.
model1.fit(
train_images,
train_labels,
epochs=5,
verbose=0,
callbacks=[tensorboard_callback, cm_callback],
validation_data=(test_images, test_labels))
The “Images” tab displays the image you just logged.

The image is scaled to a default size for easier viewing. If you want to view the unscaled original image, check “Show actual image size” at the upper left.
Related Post
- How to get the ROC curve and AUC for Keras model?
- Calculate Precision, Recall and F1 score for Keras model
- Micro and Macro Averages for imbalance multiclass classification
- Calculate F1 Macro in Keras
- PyTorch Confusion Matrix for multi-class image classification
- TensorBoard Callback of Keras with Google Colab
Run this code in Google Colab
Computes the confusion matrix from predictions and labels.
tf.math.confusion_matrix(
labels,
predictions,
num_classes=None,
weights=None,
dtype=tf.dtypes.int32,
name=None
)
Used in the notebooks
| Used in the tutorials |
|---|
|
The matrix columns represent the prediction labels and the rows represent the
real labels. The confusion matrix is always a 2-D array of shape [n, n],
where n is the number of valid labels for a given classification task. Both
prediction and labels must be 1-D arrays of the same shape in order for this
function to work.
If num_classes is None, then num_classes will be set to one plus the
maximum value in either predictions or labels. Class labels are expected to
start at 0. For example, if num_classes is 3, then the possible labels
would be [0, 1, 2].
If weights is not None, then each prediction contributes its
corresponding weight to the total value of the confusion matrix cell.
For example:
tf.math.confusion_matrix([1, 2, 4], [2, 2, 4]) ==>
[[0 0 0 0 0]
[0 0 1 0 0]
[0 0 1 0 0]
[0 0 0 0 0]
[0 0 0 0 1]]
Note that the possible labels are assumed to be [0, 1, 2, 3, 4],
resulting in a 5×5 confusion matrix.
Args |
|
|---|---|
labels
|
1-D Tensor of real labels for the classification task.
|
predictions
|
1-D Tensor of predictions for a given classification.
|
num_classes
|
The possible number of labels the classification task can have. If this value is not provided, it will be calculated using both predictions and labels array. |
weights
|
An optional Tensor whose shape matches predictions.
|
dtype
|
Data type of the confusion matrix. |
name
|
Scope name. |
Returns |
|---|
A Tensor of type dtype with shape [n, n] representing the confusionmatrix, where n is the number of possible labels in the classificationtask. |
Raises |
|
|---|---|
ValueError
|
If both predictions and labels are not 1-D vectors and have mismatched shapes, or if weights is not None and its shape doesn’tmatch predictions.
|
I am building a multiclass model with Keras.
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
model.fit(X_train, y_train, batch_size=batch_size, epochs=epochs, verbose=1, callbacks=[checkpoint], validation_data=(X_test, y_test)) # starts training
Here is how my test data looks like (it’s text data).
X_test
Out[25]:
array([[621, 139, 549, ..., 0, 0, 0],
[621, 139, 543, ..., 0, 0, 0]])
y_test
Out[26]:
array([[0, 0, 1],
[0, 1, 0]])
After generating predictions…
predictions = model.predict(X_test)
predictions
Out[27]:
array([[ 0.29071924, 0.2483743 , 0.46090645],
[ 0.29566404, 0.45295066, 0.25138539]], dtype=float32)
I did the following to get the confusion matrix.
y_pred = (predictions > 0.5)
confusion_matrix(y_test, y_pred)
Traceback (most recent call last):
File "<ipython-input-38-430e012b2078>", line 1, in <module>
confusion_matrix(y_test, y_pred)
File "/Users/abrahammathew/anaconda3/lib/python3.6/site-packages/sklearn/metrics/classification.py", line 252, in confusion_matrix
raise ValueError("%s is not supported" % y_type)
ValueError: multilabel-indicator is not supported
However, I am getting the above error.
How can I get a confusion matrix when doing a multiclass neural network in Keras?
Create a confusion matrix for neural network predictions
In this episode, we’ll demonstrate how to create a confusion matrix, which will aid us in being able to visually observe how well a neural network is predicting during inference.
We’ll continue working with the predictions we obtained from the tf.keras.Sequential model in the
last episode.
In the
last episode, we showed how to use a trained model for inference on new data in a test set it hasn’t seen before. As mentioned in that episode, we had the labels for the test set, but we didn’t
provide these labels to the network.
Additionally, we were able to see the values that the model was predicting for each of the samples in the test set by just observing the predictions themselves.
Below are the probabilities that the model assigned to whether patients from the test set were more or less likely to experience side effects from an experimental drug.
for i in predictions: print(i)
[ 0.74106830 0.25893170] [ 0.14958295 0.85041702] [ 0.96918124 0.03081879] [ 0.12985019 0.87014979] [ 0.88596725 0.11403273] ...
We then looked only at the most probable prediction for each sample.
for i in rounded_predictions: print(i)
0 1 0 1 0 ...
Although we were able to read the predictions from the model easily, we weren’t easily able to compare the predictions to the true labels for the test data.
With a confusion matrix, we’ll be able to visually observe how well the model predicts on test data.
Let’s jump into the code for how this is done.
Plotting a confusion matrix
First, we import all the required libraries we’ll be working with.
%matplotlib inline from sklearn.metrics import confusion_matrix import itertools import matplotlib.pyplot as plt
The confusion matrix we’ll be plotting comes from scikit-learn.
We then create the confusion matrix and assign it to the variable cm. T
cm = confusion_matrix(y_true=test_labels, y_pred=rounded_predictions)
To the confusion matrix, we pass in the true labels test_labels as well as the network’s predicted labels rounded_predictions for the test set.
Below, we have a function called plot_confusion_matrix() that came directly from
scikit-learn’s website. This is code that they provide in order to plot the confusion matrix.
def plot_confusion_matrix(cm, classes, normalize=False, title='Confusion matrix', cmap=plt.cm.Blues): """ This function prints and plots the confusion matrix. Normalization can be applied by setting `normalize=True`. """ plt.imshow(cm, interpolation='nearest', cmap=cmap) plt.title(title) plt.colorbar() tick_marks = np.arange(len(classes)) plt.xticks(tick_marks, classes, rotation=45) plt.yticks(tick_marks, classes) if normalize: cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis] print("Normalized confusion matrix") else: print('Confusion matrix, without normalization') print(cm) thresh = cm.max() / 2. for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])): plt.text(j, i, cm[i, j], horizontalalignment="center", color="white" if cm[i, j] > thresh else "black") plt.tight_layout() plt.ylabel('True label') plt.xlabel('Predicted label')
Next, we define the labels for the confusion matrix. In our case, the labels are titled “no side effects” and “had side effects.”
cm_plot_labels = ['no_side_effects','had_side_effects']
Lastly, we plot the confusion matrix by using the plot_confusion_matrix() function we just discussed. To this function, we pass in the confusion matrix cm and the labels
cm_plot_labels, as well as a title for the confusion matrix.
plot_confusion_matrix(cm=cm, classes=cm_plot_labels, title='Confusion Matrix')
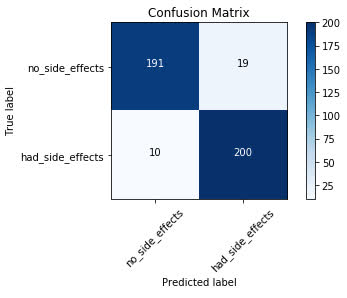
Reading a Confusion Matrix
Looking at the plot of the confusion matrix, we have the predicted labels on the x-axis and the true labels on the y-axis. The blue cells running from the top left to bottom right contain the number of samples that the model accurately predicted. The
white cells contain the number of samples that were incorrectly predicted.
There are 420 total samples in the test set. Looking at the confusion matrix, we can see that the model accurately predicted 391 out of 420 total samples. The model
incorrectly predicted 29 out of the 420.
For the samples the model got correct, we can see that it accurately predicted that the patients would experience no side effects 191 times. It incorrectly predicted that the patient would have
no side effects 10 times when the patient did actually experience side effects.
On the other side, the model accurately predicted that the patient would experience side effects 200 times that the patient did indeed experience side effects. It incorrectly predicted that
the patient would have side effects 19 times when the patient actually did not experience side effects.
As you can see, this is a good way we can visually interpret how well the model is doing at its predictions and understand where it may need some work.